Clearly there is a need for the improvement of survival rates in head and neck cancer, ideally with a reduction of severe toxicity. The most likely way to accomplish this is to better select patients for a treatment that fits their specific tumor characteristics. Currently only the clinical characteristics are used and treatment is based on site and TNM stage, which merely account for 25% of the variation in survival (211, 212, 213, 214).
The research described in this thesis aims to discover more about the individual biological tumor properties in head and neck cancer, using messenger- and microRNA data to predict which tumors will be more radioresistant and why. Eventually this could lead to a better understanding of the reasons for radiotherapy failure and an up-front adaptation of therapy (depicted in figure 1.8) to give each individual patient the best chance of survival (215, 216, 217).
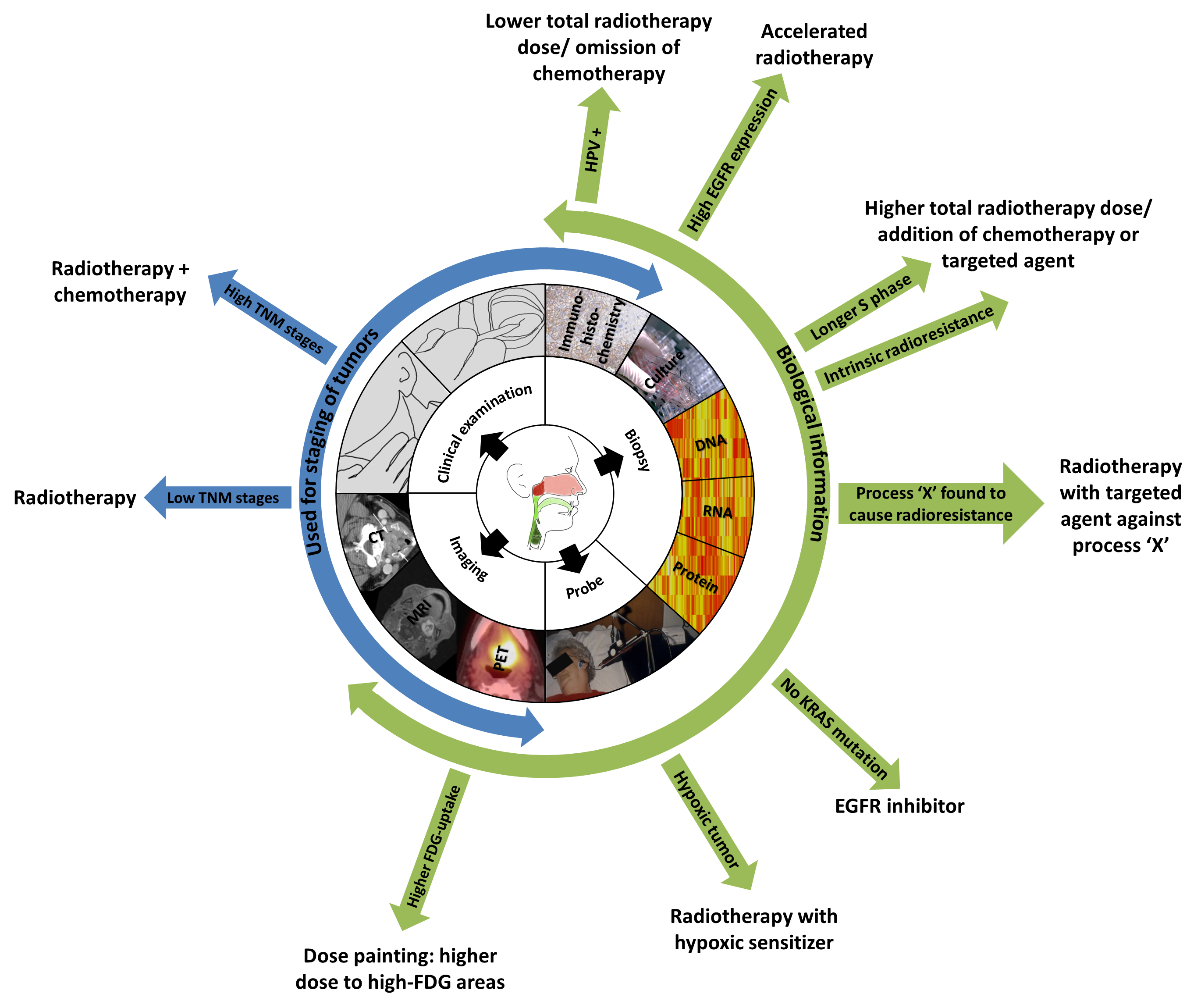
Figure 1.8. An example of the current use of clinical factors (blue) and how addition of biological knowledge could individualize and improve radiotherapy (green).
In chapter 2, we show that gene expression can improve the prediction model and adds valuable information to known clinical factors for local control after chemoradiotherapy in 75 advanced head and neck cancer patients. Chapter 3 describes the analysis of a more homogeneous series of 52 T1-2 larynx cancer patients, treated with single modality radiotherapy. Pre-treatment high expression of the putative stem cell marker CD44 correlates with local recurrence rate in this training series and in an independent validation cohort of 76 patients. Chapter 4 describes the discovery of an intrinsic radioresistance gene set on mRNA and micro RNA expression data from 32 head and neck cancer cell lines. We found that low expression of miR-203, giving more epithelial-to-mesenchymal transition, not only corresponds with intrinsic radiosensitivity, but also predicts outcome after radiotherapy in larynx cancer patients. Chapter 5 describes the comparison of published hypoxia gene sets that seem very dissimilar. However, these almost entirely different sets of genes classify 224 head and neck cancer patients nearly identically.